Molecular and Clinical Medicine
Research in this core theme is focused on understanding cellular and molecular mechanisms underpinning biological and clinical aetiology of chronic diseases (cardiovascular disease; chronic obstructive pulmonary disease (COPD); ciliary disease; diabetes; inflammatory bowel disease iron deficiency anaemia (IDA) in pregnancy) and cancer (bone, breast, brain, colorectal, lung, myeloma, pancreatic, renal, metastases) and using this knowledge to develop novel targeted therapeutic strategies.
Complementary workstreams include investigations of inflammatory and immune responses in disease; interactions with the tissue microenvironment and microbiome; establishment and validation of new, representative pre-clinical disease models; and development of enhanced clinical treatment pathways.
Staff list
- Professor Angel Armesilla (Professor of Cardiovascular Molecular Pharmacology)
- Dr Kesley Attridge (Senior Lecturer in Biomedical Science)
- Dr Paul Barrow (Senior Lecturer in Biomedical Science)
- Professor Supratik Basu (Professor of Haematology)
- Professor Matthew Brookes (Professor of Gastroenterology)
- Dr Matthew Conner (Senior Lecturer in Biology)
- Professor James Cotton (Professor of Cardiology)
- Professor David Churchill (Professor of Obstetrics)
- Dr Janine Fletcher (Senior Lecturer in Human and Clinical Physiology)
- Professor Rousseau Gama (Professor of Laboratory and Metabolic Medicine)
- Dr Paraskevi Goggolidou (Reader in Molecular Genetics)
- Dr Aikaterina Karakoula (Senior Research Fellow)
- Professor Paul Kirkham (Professor of Cell Biology)
- Dr Mark Morris (Director of RIHS; Reader in Molecular Oncology)
- Dr Iain Nicholl (Senior Lecturer in Biomedical Science)
- Dr Elizabeth O’Gara (Principal Lecturer in Employability)
- Dr Opeolu Ojo (Senior Lecturer in Biotechnology)
- Dr Hafid Omar (Senior Lecturer in Cancer Research)
- Dr Mitesh Patel (Senior Lecturer in Physiology)
- Professor Thillagavathie Pillay (Professor of Neonatology)
- Professor Baldev Singh (Professor of Diabetes)
- Professor Helen Steed (Professor of Gastroenterology and Medical Education)
- Dr James Vickers (Senior Lecturer in Biomedical Science)
- Professor Weiguang Wang (Professor of Experimental Cancer Therapeutics)
- Professor Tracy Warr (Associate Dean Research and Knowledge Exchange; Professor of Neuro-Oncology)
Examples of Current Projects
PI: Dr Mark Morris
As cancers develop, they can acquire the ability to move through the body and continue to grow in organs or tissue that is distant to the originating tissue, a process that is termed metastasis. When breast cancer is not successfully treated, a common site of metastasis is the brain. The breast cancer metastasis group, led by Dr Mark Morris, is investigating the molecular switches that can determine the ability of breast cancer cells to travel to and then grow in the brain. Specifically, this long-term collaborative project is investigating epigenetic changes and gene mutations found in breast cancer cells that metastasise to the brain.
The key objectives of this research are to identify molecular markers of brain metastasis risk and to identify new cellular pathways that can be targeted as therapies against brain metastasis.
This research is ongoing and a collaboration between Dr Morris’s group, and the research groups of Professor Tracy Warr, Professor Angel Armesilla and Professor Weiguang Wang.
Publications:
- Pangeni RP, Olivaries I, Huen D, Buzatto VC, Dawson TP, Ashton KM, Davis C, Brodbelt AR, Jenkinson M, Bièche I, Yang L, Latif F, Darling JL, Armesilla A, Warr TJ, Morris MR. Genome-wide methylation analyses identifies genes dysregulated in breast tumors that metastasise to the brain. (in preparation,2021)
- Morris MR, I Olivares, T Dawson, K Ashton, C Davis, M Jenkinson, A Brodbelt, A Armesilla, T Warr. Identifying breast-to-brain metastasis-associated gene mutations by whole exome sequencing. Springer Nature https://wlv.openrepository.com/bitstream/handle/2436/623120/Abstract%20Morris%20et%20al%202018%20.pdf?sequence=3
- Olivares I, Pangeni RP, Huen D, Dawson TP, Ashton K, Davis CHG, Jenkinson MD, Brodbelt AR, Wang W, Darling JW, Warr TJ, Morris MR. (2018) Identifying epigenetic changes in breast tumours that metastasise to the brain. European Journal of Surgical Oncology 44, S36. https://www.ejso.com/article/S0748-7983(18)30628-0/abstract
- Pangeni RP, Channathodiyil P, Huen DS, Eagles LW, Johal BK, Pasha D, Hadjistephanou N, Nevell O, Davies CL, Adewumi AI, Khanom H, Samra IS, Buzatto VC, Chandrasekaran P, Shinawi T, Dawson TP, Ashton KM, Davis C, Brodbelt AR, Jenkinson MD, Bièche I, Latif F, Darling JL, Warr TJ, Morris MR. (2015). The GALNT9, BNC1 and CCDC8 genes are frequently epigenetically dysregulated in breast tumours that metastasise to the brain. Clin Epigenetics 7 (1), 57 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4457099/
Media:
- https://www.ncri.org.uk/gene-switches-could-predict-when-breast-cancers-will-spread-to-the-brain/
- https://www.itv.com/news/central/2014-11-05/bbb
- https://www.expressandstar.com/news/2014/11/06/university-of-wolverhampton-researchers-developing-breast-cancer-blood-test/
- https://www.sciencedaily.com/releases/2014/11/141104194623.htm
The immune system is capable of mounting robust anti-tumour immune responses, but in glioblastoma (the most malignant type of adult brain tumours), these responses are heavily suppressed. If this immunosuppression could be overcome, patients with glioblastoma would have a potent mechanism for fighting their cancer from within. To make this happen, we need to know:
- Which immune cells are present within GBM tumours? This will identify the cells we need to focus on protecting from immunosuppression. Their location within the tumour makes them ideal candidates for therapy, as they are well placed to attack the tumour once they are restored to full function.
We use confocal microscopy to image brain tumour-infiltrating T cells, B cells, macrophages and microglia, and to explore their interactions with the tumour and each other. The work is undertaken by PhD student Rye McIntyre-Bhatty and funded by the Colin Oliphant Charitable Trust. - Which proteins are used by GBM tumours to suppress the immune response? GBM tumours produce a huge array of proteins that are released into their surrounding environment (at last count, our laboratory has identified more than 2,500 such proteins!), but only a fraction of these are responsible for immunosuppression. Identifying these proteins will lead to new therapies to block them, so that the anti-tumour immune response is restored.
We use mass spectrometry and a primary human T cell screening platform to identify candidate proteins for future therapeutic targeting studies. The work is undertaken by Rye McIntyre-Bhatty and Ourania Gkanatsiou in collaboration with the Devitt laboratory at Aston University, and is funded by Headcase Cancer Trust.
Both of these projects are made possible by an ongoing collaboration with Tracy Warr’s group, and the Brain Tumour North West and the Walton Research Tissue Banks.
One of the research lines in the laboratory of Professor Angel Armesilla in RIHS, investigates the molecular mechanisms that control blood vessel formation. The formation of new blood vessels from pre-existing ones (called angiogenesis) is a tightly regulated process essential for tissue and organ development. De-regulation of this process results in excessive or insufficient formation of blood vessels leading to severe human pathologies. For example, solid tumors promote the formation of new blood vessels that perfuse the tumor and deliver oxygen and nutrients so the tumoral cells can grow. If we could stop the formation of these new blood vessels, we could kill the tumor by “starvation”. In other cases, blockage of blood vessels restricts oxygen delivery to an organ, for example the obstruction of a coronary artery during a heart attack. In this case, the formation of new blood vessels would allow to bypass the blocked area and to supply oxygen to the ischaemic heart.
We are characterizing the molecular mechanisms that control the formation of new blood vessels in healthy and pathological situations. We will use this information to design novel therapeutic interventions that stop (in cancer) or promote (after myocardial infarction) the formation of blood vessels.
Recent publications by the group in this area:
- Njegic A et al (2021) J Mol Cell Cardiol. 156:79-81
- Savage AM et al. (2019) Nat Commun. 10(1):732
- Kurusamy S et al (2017) J Mol Cell Cardiol. 109:38-47
- Baggott RR et al (2014) Arterioscler Thromb Vasc Biol. 34(10):2310-20
For more information, please go to the webpage for Professor Armesilla’s group at Armesillalab.com.
Cardiovascular disease (CVD) encompasses a number of different conditions including coronary heart disease, stroke, and peripheral vascular disease. CVD currently represents a major cause of death, morbidity, and economic burden all around the world, accounting for more than 17 million deaths each year. In the UK, more than a quarter of all deaths are caused by CVD; that is more than 160,000 deaths each year – or one death every three minutes.
Our team is trying to identify key molecules in the progression of CVD. PhD student Kinza Khan is studying the pathological mechanisms occurring in diseased arteries during atherosclerosis, aneurysm, and peripheral vascular disease. These investigations are being carried out in close collaboration with Prof James Cotton from Wolverhampton New Cross Hospital who leads the clinical part of the projects. Recently, vascular surgeons Dr Mike Wall and Dr Andrew Garnham from Russells Hall Hospital are also actively collaborating in our investigations on peripheral arterial disease in diabetic patients, a disease that, unfortunately, in many cases leads to leg amputation.
Also, studying arterial disease, PhD student Jude Ihguda is almost finishing his PhD on the role of PMCA, a calcium transporter protein, in pulmonary arterial hypertension.
To conduct these projects, we use a combination of experimental approaches involving transcriptomics, proteomics, and molecular and cellular analysis using human endothelial cells isolated from aortic or pulmonary artery.
For more information, please go to the webpage for Professor Armesilla’s group at Armesillalab.com.
Glioblastoma is the most common primary malignant tumour of the brain affecting the elderly, with a rising incidence in the UK. At present there is no cure for patients suffering from glioblastoma.
These tumours are characterised by the abundance of microvascular proliferation and invasive glioma cells, which accounts for its poor prognosis. In humans, vascular endothelial growth factor-A (VEGF-A) has been implicated in glioblastoma vascularisation. VEGF-A antagonists are being used in clinic to treat glioblastoma. However, the effect of these agents is short-lived, with limited impact on overall survival.
After a brief period of tumor reduction, the tumoral cells acquire new mechanisms to evade the treatment and the tumor becomes revascularised, promoting a new wave of tumour growth and invasiveness.
As a first step towards understanding the molecular mechanisms implicated in anti-VEGF-A evasion, PhD student Miebaka Ian-Gobo is studying the tumoral response to anti-VEGF therapy using primary human cells isolated from brain tumor patients. She is treating these cells with anti-VEGF agents to determine their evasion response.
High vascularisation in glioblastoma tumors is the consequence of high levels of hypoxia. PhD student Shervin Najafi is investigating in our laboratory the molecular changes associated to hypoxia in human glioblastoma cells. These investigations will shed new insights into the response of glioblastoma cells to a hypoxic environment and will reveal new molecular targets to design new therapeutic interventions.
These projects are carried out in close collaboration with members of the Brain Tumor Group in RIHS, Dr Aikaterine Karakoula and Professor Tracy Warr.
For more information, please go to the webpage for Professor Armesilla’s group at Armesillalab.com.
PI: Dr Alice Clark
This diverse protein family punch holes in their target cell membrane, altering membrane permeability. The family is found in all kingdoms of life and act as virulence factors (allowing bacteria to infect patients), self-defence (for e.g. in the patient’s immune system) or compartment release. Typically, these proteins are produced as water-soluble monomers and undergo a large conformational change during membrane insertion. We study members of the small beta-pore forming proteins (β-PFTs), so called because the pore that spans the membrane is comprised of beta strands forming a circular beta-barrel.
The latest structure solved by this group of researchers is available to visualise and explore in 3D here.
Lead by Dr Alice Clark, this project aims to solve structures of this protein family using CryoEM methodology. We also collaborate with University of Leicester, and work closely with Dr Christos Savva. The data is collected on the Titan Krios microscope within the Midlands Regional Cryo-Electron Microscope Facility.
For more information about projects, collaborations or opportunities to join this group email Dr. Alice Clark at alice.clark@wlv.ac.uk.
Relevant Publications
- Savva et al., The pore structure of Clostridium perfringens epsilon toxin. Nat Commun. 2019 https://doi.org/10.1038/s41467-019-10645-8 )
PI: Dr Alice Clark
Ageing in humans can be thought of as the accumulation of molecular and cellular changes over time and is a known risk factor for many human diseases. The ageing-associated proteopathy diseases occur when certain proteins, within cells, become structurally abnormal and misfolded, which disrupts the function of cells and resulting tissues. Proteopathy diseases include Alzheimer’s disease, Parkinson’s disease, and type II diabetes, which all result in protein aggregation into amyloid fibres within cells. Increasing molecular chaperones such as small heat shock proteins (sHSP) in the diseased tissue can suppress protein aggregation, and therefore the associated toxicity in numerous animal and cellular models of these diseases. A key sHSP chaperone, αB-crystallin, maintains the cellular environment, preventing further aggregation and amyloid fibre formation. In the eye lens, αB crystallin is involved in maintaining transparency and correct optical properties and plays a protective role within the retina. In the brain it also plays a protective role,and is upregulated in several neurodegenerative diseases. However, very little is known about the precise molecular details of how it does this. Solving structures of sHSPs will reveal the molecular details, showing us what they do, both to maintain healthy tissues, and in diseased tissues such as those found in cataracts and Alzheimer’s disease.
Led by Dr Alice Clark this project aims to determine the structure of sHSP proteins, in particular how determining the proteins structure can help us further understand the function and mechanism in plays within the cell. We use both x-ray crystallography and cryo-electron microscopy to study the structures of proteins targets.
This project is currently funded by the Lord Paul Fellowship.
For more information about projects, collaborations or opportunities to join this group email Dr. Alice Clark at alice.clark@wlv.ac.uk.
Relevant Publications
- Clark et al. Terminal Regions Confer Plasticity to the Tetrameric Assembly of Human HspB2 and HspB3. J. Mol. Biol. 2018. https://doi.org/10.1016/j.jmb.2018.06.047
- Slingsby and Clark. Book chapter in “Structure and action of Molecular Chaperones”. Series in Structural Biology: Volume 6. 2016. Chapter 4: The family of sHSP assembly and binding functions. https://doi.org/10.1142/9789814749336_0004
- Clark et al. sHSP in the eye lens: crystallin mutations, cataract and proteostasis. Int. J. Biochem. Cell Biol. 2012. https://doi.org/10.1016/j.biocel.2012.02.015
- Slingsby and Clark. Breaking down order to keep cells tidy. Chem. Biol. 2012. https://doi.org/10.1016/j.chembiol.2012.05.003
- Clark et al. Crystal structure of R120G disease mutant of human αB-crystallin domain dimer shows closure of a groove. J. Mol. Biol. 2011. https://doi.org/10.1016/j.jmb.2011.02.020
Cell plasticity plays an important role in immunity. Defects in this important cellular process can lead to failures in immune–mediated repair and an inability to resolve the inflammatory response due to a detrimental impact on phagocytosis and a failure to clear apoptotic cells or bacteria. This can lead to a state of chronic inflammation, which is a characteristic of several mucosal inflammatory diseases including Chronic Obstructive Pulmonary Disease (COPD) and Crohn’s Disease, both of which are marked by oxidative stress. Determinants of the microenvironment, in which inflammatory cells exist, include the presence of PAMPs, DAMPS, and oxidative stress. Recently, DAMPS such as Heat Shock Protein 70, have been shown to modulate the inflammatory response by promoting an anti-inflammatory (M2) macrophage response which possesses increased phagocytic activity to help remove damaged cells and debris. In contrast, preliminary experiments in our lab demonstrated that when HSP70 is modified by oxidative stress, it can promote a more damaging pro-inflammatory (M1) phenotype with reduced capacity to phagocytose. In COPD, a disease driven by oxidative stress, macrophages have been shown to behave in a more pro-inflammatory (M1) phenotype. Furthermore, extracellular levels of HSP70 have also been shown to be significantly higher in tissues exposed to chronic inflammation, such as that seen in COPD lungs. This project, led by Professor Kirkham and his PhD student (Reham Hassanan), will assess whether HSP70 modified by continuous exposure to oxidative stress, which is present in COPD, drives the formation of a pro-inflammatory M1 phenotype and the mechanism by which it achieves this. Moreover, compounds that target the molecular pathways involved in driving this M1 phenotype will be examined for their ability to prevent or even reverse this switch to an M2 phenotype, thereby identifying potential novel treatment strategies for COPD.
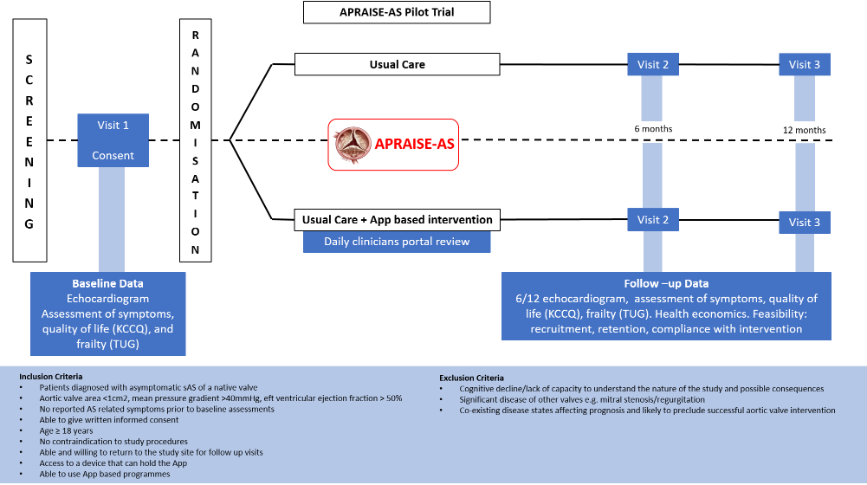
A Pilot Study to assess the safety, effectiveness and practical utility of remote patient monitoring to guide the timing of valve intervention in patients with asymptomatic severe aortic valve stenosis (APRAISE AS).
Aortic stenosis (AS) is the commonest presentation of heart valve disease seen in clinical practice affecting predominantly older adults. It is associated with poor prognosis once severe and patients are symptomatic, but can be effectively treated with an aortic valve replacement procedure. However, intervention at the time of symptom development is challenging since the asymptomatic latent period is unpredictable and patients experience rapid deterioration and potentially irreversible myocardial damage on symptom development, leading to poor outcomes.
Our study will determine how efficiently digital technology can be embedded into the clinical management pathway for patients diagnosed with asymptomatic severe AS (sAS). We will assess the utility of a novel digital application to support remote patient monitoring coupled with near real-time clinician surveillance to facilitate assessment of symptom development, health-related quality of life (assessed using the Kansas City Cardiomyopathy Questionnaire (KCCQ)) and functional capacity (as a marker of frailty) to pre-empt potential deterioration and guide the optimal timing of valve replacement.
The app/portal will supplement a routine “watchful waiting” approach consisting of a 6 monthly out-patient clinic review. Although watchful waiting is the standard of care, there are limitations associated with this strategy in that we rely on patients to self-report symptoms as and when they occur. Clinical deterioration and possible worsening of disease severity may be overlooked or underestimated resulting in poor clinical outcomes in the longer term.
Our study will also help to inform the selection of the most appropriate outcome measures for a full-scale randomised controlled trial.
Seed corn funding was gained from the West Midlands Clinical Research Network, and funding is being sort with an application to NIHR RfPB currently under review. The Study has also been supported by a generous donation from the Rotha Abraham bequest.
The study has been developed in collaboration with the West Midland Academic Health Science Network, the Birmingham Clinical Trials Network and the Smartphone App has been developed in collaboration with local software development firm, B13.
The study is led by Professor James Cotton, University of Wolverhampton, and Dr Nazish Khan, Royal Wolverhampton NHS Trust.
The study is currently in the set up phase.
Fibrosis is an important process for tissue repair following injury. However, should this process become uncoordinated and overactive, it can lead to tissue remodelling and disease. This is a serious medical problem and is associated with several chronic diseases linked to specific organs, such as the kidney, lung, heart and liver. Many of these conditions have a poor prognosis and eventually result in death, and treatment options are currently limited with a high unmet medical need for new therapeutic intervention strategies. In order to meet this challenge, key to achieving this goal is to gain a greater understanding of, and validate, the cellular and molecular mechanisms that operate in each of these conditions. One of the mechanisms driving fibrosis involves dysfunctional mitochondria that fail to be removed by mitophagy following damage by continues exposure to oxidative stress, a key aetiological factor in disease pathogenesis.
To this end, the University in association with its partner, the Royal Wolverhampton NHS Trust, has entered into a collaboration with a biotechnology company to fund a postdoctoral research programme to evaluate the effectiveness of novel drugs in restoring mitophagy and mitochondrial function, thereby preventing/halting the fibrotic response. Leading this effort are Professor Paul Kirkham (University of Wolverhampton), Dr Ahmed Fahim (Head of Respiratory & Fibrosis clinic, New Cross Hospital) and Dr Lawrence Eagles (University of Wolverhampton).
Our work focuses on understanding the molecular mechanisms of Autosomal Recessive Polycystic Kidney Disease (ARPKD), a rare paediatric disease for which no pharmacological treatment currently exists. ARPKD is a genetic disorder affecting ~1:20,000 and is a common cause of perinatal death. It manifests as extreme bilateral enlargement of cystic kidneys in utero, associated with hepatic ductal plate abnormalities and pulmonary hypoplasia. In those patients who survive the perinatal period, the majority will require renal replacement therapy (kidney dialysis or transplantation) within the first decade of life. Recently, however, ARPKD patients have been diagnosed in their 30s with relatively mild renal insufficiency, suggesting a previously unrecognised, wide spectrum of disease severity.
At the molecular level, Polycystic Kidney and Hepatic Disease 1 (PKHD1) and DAZ Interacting Zinc Finger Protein 1 Like (DZIP1L) mutations are associated with ARPKD. DZIP1L mutations are involved in a rarer subset of patients featuring a moderate ARPKD presentation. PKHD1 encodes the cilia localising protein Fibrocystin and DZIP1L encodes a protein that localises to the ciliary diffusion barrier. The range of disease severity observed in ARPKD may be due to the various types of mutations observed in these two genes, however, it remains possible that additional modifier genes may play a role in ARPKD manifestation and severity. We have shown that one such potential gene, ATMIN, plays a role in kidney morphogenesis via modulation of the Wnt signalling pathway (Goggolidou et al, 2014; Richards et al, 2019).
This research project seeks to investigate the role of Planar Cell Polarity (PCP) signalling in human ARPKD cell lines and tissues; as well as characterise the changes in network interactions between ATMIN, Fibrocystin and related proteins in both normal and ARPKD human kidney. This is to identify whether ATMIN is a modifier gene in ARPKD and may be of potential value as a biomarker of disease severity, through its interactions with Fibrocystin.
To this end we are using DNA-seq and RNA-seq approaches in ARPKD cells and tissues to identify the role of various mutations in ARPKD and dissect the complex transcriptional work of fibrocystin (collaboration with Prof Patricia Wilson, UCL). We are also employing ChIP-sequencing techniques to dissect the DNA-protein interactions of ATMIN (collaboration with Prof Shona Murphy, Oxford University). Our work, conducted by Taylor Richards in his PhD studies is funded by the PKD charity UK and is hoping to shed light on the complex molecular network of ARPKD.
https://www.sciencedirect.com/science/article/pii/S0925443918304460?via%3Dihub
https://academic.oup.com/hmg/article/23/20/5303/2900648
PI details: Paraskevi Goggolidou, BSc, DPhil, FHEA, FIBMS
Reader in Molecular Genetics
Faculty of Science and Engineering
University of Wolverhampton
Tel: +44 (0)1902 3211552
Email:p.goggolidou@wlv.ac.uk
Twitter: @PGoggolidou


/prod01/wlvacuk/media/departments/digital-content-and-communications/images-18-19/iStock-163641275.jpg)
/prod01/wlvacuk/media/departments/digital-content-and-communications/images-2024/250630-SciFest-1-group-photo-resized-800x450.png)
/prod01/wlvacuk/media/departments/digital-content-and-communications/images-18-19/210818-Iza-and-Mattia-Resized.jpg)
/prod01/wlvacuk/media/departments/digital-content-and-communications/images/Maria-Serria-(teaser-image).jpg)
/prod01/wlvacuk/media/departments/digital-content-and-communications/images-2024/241014-Cyber4ME-Project-Resized.jpg)
/prod01/wlvacuk/media/departments/digital-content-and-communications/images-18-19/210705-bric_LAND_ATTIC_v2_resized.jpg)